|
|
| C4orf32, C4orf34, and C4orf40 |
Novel Peptides derived from Chromosome 4 Proteins |
C4orf32:
AIMS/HYPOTHESIS:
Recent genome-wide association studies performed in selected patients and control participants have provided strong support for several new type 2 diabetes susceptibility loci. To get a better estimation of the true risk conferred by these novel loci, we tested a completely unselected population of type 2 diabetes patients from a Norwegian health survey (the HUNT study).
METHODS:
We genotyped single nucleotide polymorphisms (SNPs) in PKN2, IGFBP2, FLJ39370 (also known as C4ORF32), CDKAL1, SLC30A8, CDKN2B, HHEX and FTO using a Norwegian population-based sample of 1,638 patients with type 2 diabetes and 1,858 non-diabetic control participants (the HUNT Study), for all of whom data on BMI, WHR, cholesterol and triacylglycerol levels were available. We used diabetes, measures of obesity and lipid values as phenotypes in case-control and quantitative association study designs.
RESULTS:
We replicated the association with type 2 diabetes for rs10811661 in the vicinity of CDKN2B (OR 1.20, 95% CI: 1.06-1.37, p=0.004), rs9939609 in FTO (OR 1.14, 95% CI: 1.04-1.25, p=0.006) and rs13266634 in SLC30A8 (OR 1.20, 95% CI: 1.09-1.33, p=3.9 x 10(-4)). We found borderline significant association for the IGFBP2 SNP rs4402960 (OR 1.10, 95% CI: 0.99-1.22). Results for the HHEX SNP (rs1111875) and the CDKAL1 SNP (rs7756992) were non-significant, but the magnitude of effect was similar to previous estimates. We found no support for an association with the less consistently replicated FLJ39370 or PKN2 SNPs. In agreement with previous studies, FTO was most strongly associated with BMI (p=8.4 x 10(-4)).
CONCLUSIONS/INTERPRETATION:
Our data show that SNPs near IGFBP2, CDKAL1, SLC30A8, CDKN2B, HHEX and FTO are also associated with diabetes in non-selected patients with type 2 diabetes.
Jun MH, Jun YW, Kim KH et al., BMB Rep. 2014 Feb 6. pii: 2576. [Epub ahead of print]
Obesity is an established risk factor for type 2 diabetes (T2D) and they are metabolically related through the mechanism of insulin resistance. In order to explore how common genetic variants associated with T2D correlate with body mass index (BMI), we examined the influence of 25 T2D associated loci on obesity risk. We used 5056 individuals (2528 sib-pairs) recruited in Indian Migration Study and conducted within sib-pair analysis for six obesity phenotypes. We found associations of variants in CXCR4 (rs932206) and HHEX (rs5015480) with higher body mass index (BMI) (β=0.13, p=0.001) and (β=0.09, p=0.002), respectively and weight (β=0.13, p=0.001) and (β=0.09, p=0.001), respectively. CXCR4 variant was also strongly associated with body fat (β=0.10, p=0.0004). In addition, we demonstrated associations of CXCR4 and HHEX with overweight/obesity (OR=1.6, p=0.003) and (OR=1.4, p=0.002), respectively, in 1333 sib-pairs (2666 individuals). We observed marginal evidence of associations between variants at six loci (TCF7L2, NGN3, FOXA2, LOC646279, FLJ39370 and THADA) and waist hip ratio (WHR), BMI and/or overweight which needs to be validated in larger set of samples. All the above findings were independent of daily energy consumption and physical activity level. The risk score estimates based on eight significant loci (including nominal associations) showed associations with WHR and body fat which were independent of BMI. In summary, we establish the role of T2D associated loci in influencing the measures of obesity in Indian population, suggesting common underlying pathophysiology across populations.
Gupta V, Vinay DG, Sovio U et al., PLoS One. 2013;8(1):e53944. doi: 10.1371/journal.pone.0053944. Epub 2013 Jan 17.
OBJECTIVE:
Several whole-genome association studies have reported identification of type 2 diabetes susceptibility genes in various European-derived study populations. Little investigation of these loci has been reported in other ethnic groups, specifically African Americans. Striking differences exist between these populations, suggesting they may not share identical genetic risk factors. Our objective was to examine the influence of type 2 diabetes genes identified in whole-genome association studies in a large African American case-control population.
RESEARCH DESIGN AND METHODS:
Single nucleotide polymorphisms (SNPs) in 12 loci (e.g., TCF7L2, IDE/KIF11/HHEX, SLC30A8, CDKAL1, PKN2, IGF2BP2, FLJ39370, and EXT2/ALX4) associated with type 2 diabetes in European-derived populations were genotyped in 993 African American type 2 diabetic and 1,054 African American control subjects. Additionally, 68 ancestry-informative markers were genotyped to account for the impact of admixture on association results.
RESULTS:
Little evidence of association was observed between SNPs, with the exception of those in TCF7L2, and type 2 diabetes in African Americans. One TCF7L2 SNP (rs7903146) showed compelling evidence of association with type 2 diabetes (admixture-adjusted additive P [P(a)] = 1.59 x 10(-6)). Only the intragenic SNP on 11p12 (rs9300039, dominant P [P(d)] = 0.029) was also associated with type 2 diabetes after admixture adjustments. Interestingly, four of the SNPs are monomorphic in the Yoruba population of the HAPMAP project, with only the risk allele from the populations of European descent present.
CONCLUSIONS:
Results suggest that these variants do not significantly contribute to interindividual susceptibility to type 2 diabetes in African Americans. Consequently, genes contributing to type 2 diabetes in African Americans may, in part, be different from those in European-derived study populations. High frequency of risk alleles in several of these genes may, however, contribute to the increased prevalence of type 2 diabetes in African Americans.
Lewis JP, Palmer ND, Hicks PJ et al., Diabetes. 2008 Aug;57(8):2220-5. doi: 10.2337/db07-1319. Epub 2008 Apr 28.
C4orf32 & C4orf34:
Human genome projects have enabled whole genome mapping and improved our understanding of the genes in humans. However, many unknown genes remain to be functionally characterized. In this study, we characterized human chromosome 4 open reading frame 34 gene (hC4orf34). hC4orf34 was highly conserved from invertebrate to mammalian cells and ubiquitously expressed in the organs of mice, including the heart and brain. Interestingly, hC4orf34 is a novel ER-resident, type I transmembrane protein. Mutant analysis showed that the transmembrane domain (TMD) of hC4orf34 was involved in ER retention. Overall, our results indicate that hC4orf34 is an ER-resident type I transmembrane protein, and might play a role in ER functions including Ca2+ homeostasis and ER stress.
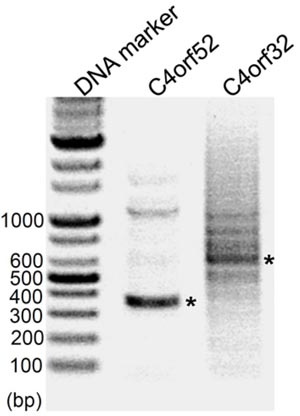
|
Each PCR band (*) was confirmed as respectively, using T-A cloning and sequencing. |
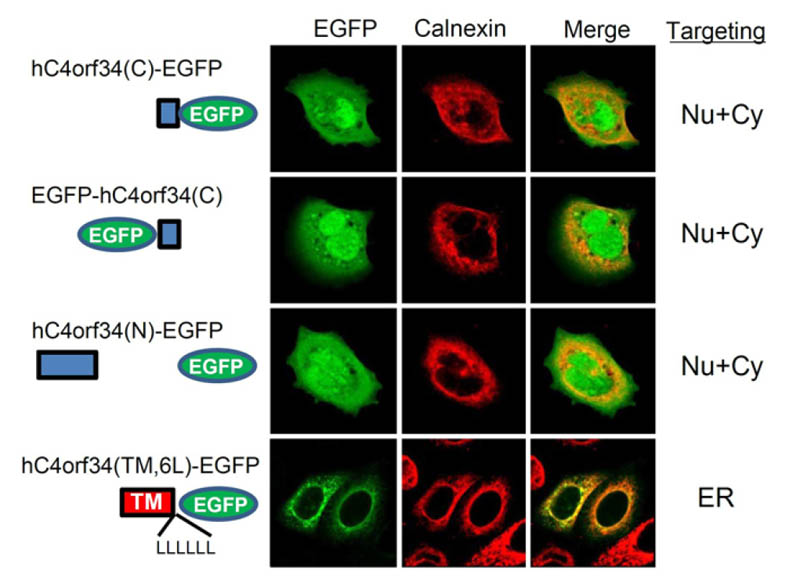
hC4orf34 and mutants were transfected into HEK293T cells. Several hC4orf34 constructs were co-localized with calnexin, but hC4orf34©-GFP and hC4orf34(N) were not, suggesting that TMD of hC4orf34 might be involved in ER targeting. (ER, endoplasmic reticulum; Cy, cytosol; Nu, nuclear)
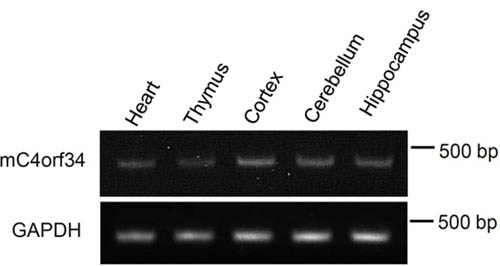
RT-PCR revealed that C4orf34 mRNA was expressed in tissues including heart, thymus, brain cortex, hippocampus and cerebellum.
Jun MH, Jun YW, Kim KH, Lee JA, Jang DJ, BMB Rep. 2014 Feb 6. pii: 2576. [Epub ahead of print]
C4orf40:
Proteomes are characterized by large protein-abundance differences, cell-type- and time-dependent expression patterns and post-translational modifications, all of which carry biological information that is not accessible by genomics or transcriptomics. Here we present a mass-spectrometry-based draft of the human proteome and a public, high-performance, in-memory database for real-time analysis of terabytes of big data, called ProteomicsDB. The information assembled from human tissues, cell lines and body fluids enabled estimation of the size of the protein-coding genome, and identified organ-specific proteins and a large number of translated lincRNAs (long intergenic non-coding RNAs). Analysis of messenger RNA and protein-expression profiles of human tissues revealed conserved control of protein abundance, and integration of drug-sensitivity data enabled the identification of proteins predicting resistance or sensitivity. The proteome profiles also hold considerable promise for analysing the composition and stoichiometry of protein complexes. ProteomicsDB thus enables navigation of proteomes, provides biological insight and fosters the development of proteomic technology.
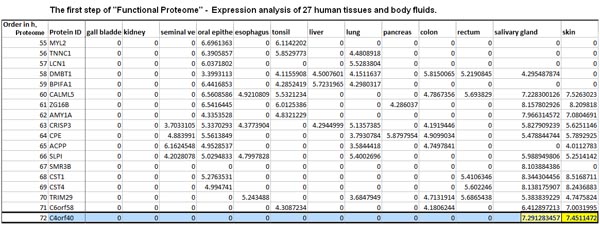
Wilhelm M, Schlegl J, Hahne H et al., Nature. 2014 May 29;509(7502):582-7. doi: 10.1038/nature13319.
Saliva contains a large number of proteins that participate in the protection of oral tissue. Exosomes are small vesicles (30-100 nm in diameter) with an endosome-derived limiting membrane that are secreted by a diverse range of cell types. We have recently demonstrated that exosomes are present in human whole saliva. In this study, we found that whole saliva contained at least two types of exosomes (exosome I and exosome II) that are different in size and protein composition. Proteomic analysis revealed that both types of exosomes contained Alix, Tsg101 and Hsp70, all exosomal markers, immunoglobulin A and polymeric immunoglobulin receptor, whereas they had different protein compositions. Most of dipeptidyl peptidase IV known as CD26 in whole saliva, was present on the exosome II and metabolically active in cleaving chemokines (CXCL11 and CXCL12). Human whole saliva exosomes might participate in the catabolism of bioactive peptides and play a regulatory role in local immune defense in the oral cavity.
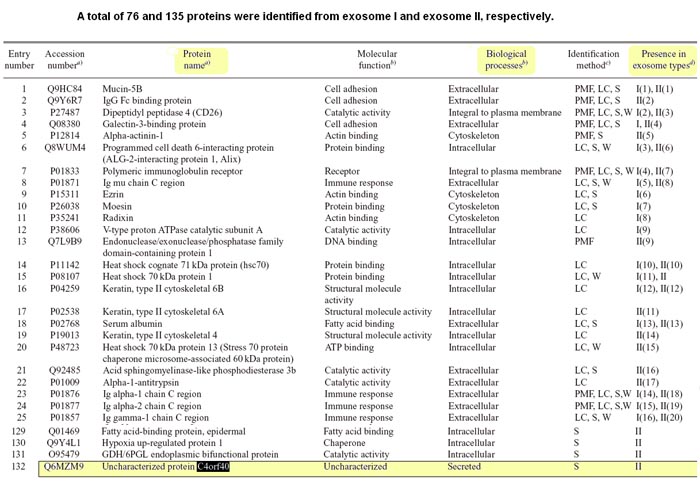
Ogawa Y, Miura Y, Harazono A et al., Biol Pharm Bull. 2011;34(1):13-23.
|
|
|
%C4orf32%;%C4orf34%;%C4orf40%
|
|
|


