|
|
|
Neuropeptide S
(NPS)
|
A novel modulator of arousal and
possibly anxiety-related behavior |
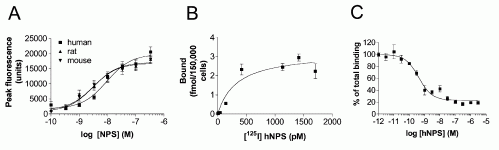
Pharmacological Characterization of the Human NPS Receptor.
(A) Dose response curve of [Ca2+]i mobilization induced by human,
rat, and mouse NPS in an HEK cell line stably expressing human NPS
receptor. (B) Saturation binding of [125I] Tyr10-NPS (4
pM to 1.7 nM) to CHO cells stably expressing human NPS receptor. (C)
Displacement of 0.15 nM [125I] Y10-NPS by increasing concentrations
of unlabeled human NPS. Data from triplicate experiments are shown
as means ± SEM.
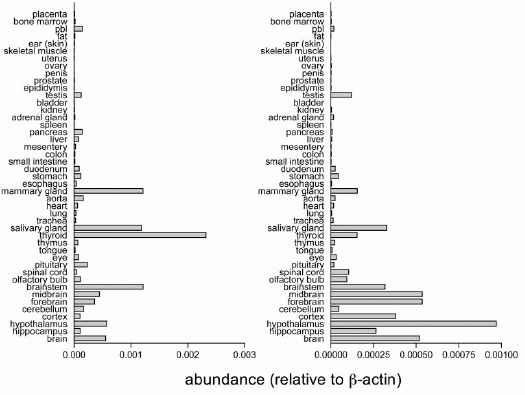
Tissue Distribution of NPS Precursor and NPS Receptor mRNA in
Rat Tissues. Quantitative RT-PCR was used to measure transcript
levels of NPS precursor (left) and NPS receptor mRNA (right) in 45
rat tissues. Transcript levels were normalized to ?actin. pbl,
peripheral blood leucocytes. 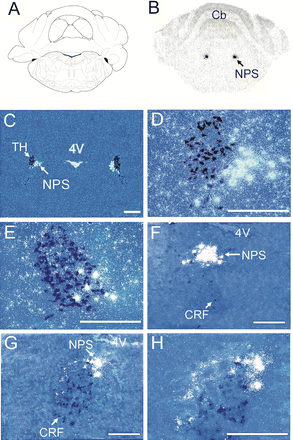
Expression of NPS Precursor mRNA in the Pontine Area of the
Rat Brain. (A) Schematic drawing of the section shown in (B). The
level is at bregma -9.80 mm (Paxinos and Watson, 1997, reprinted
with permission from Elsevier). (B) Representative autoradiogram of
NPS mRNA expression in LC area. (C–E) Dark-field images of double in
situ hybridization of NPS precursor mRNA (white) and TH mRNA (dark
blue) in LC area. (D) Higher magnification of the area indicated by
an arrow in (C). (E) Higher magnification of a more caudal section.
(F–H) Dark-field images of double in situ hybridization of NPS
precursor mRNA (white) and CRF mRNA (dark blue) at mid-level of LC
area (F) and rostral LC (G). (H) Higher magnification of the area
indicated by an arrow in (G). TH, tyrosine hydroxylase; NPS,
neuropeptide S; CRF, corticotropin-releasing factor. Landmarks: Cb,
cerebellum; 4V, fourth ventricle. Scale bar, 500 µm in (C), 250 µm
in all other pictures.
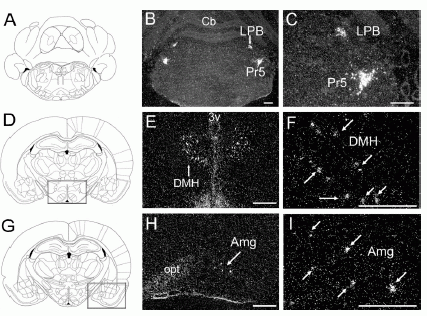
Distribution of NPS Precursor mRNA Expression in Rat Brain.
(A, D, and G) Drawings of the sections illustrated in (B) and (C)
(Bregma -9.68 mm), (E) and (F) (Bregma -2.80 mm), and (H) and (I)
(Bregma -3.14 mm), respectively (Paxinos and Watson, 1997). (B, C,
E, F, H, and I) Dark-field images of NPS precursor mRNA expression
in coronal sections of rat brain. (E and H) Expression of NPS
precursor mRNA in boxed regions in (D) and (G), respectively. (C, F,
and I) Higher magnification of the area indicated by an arrow in
(B), (E), and (H), respectively. Arrows in (F) and (I) indicate
single cells showing hybridization signals for NPS precursor mRNA.
LPB, lateral parabrachial nucleus; Pr5, principle sensory 5 nucleus;
DMH, dorsomedial hypothalamic nucleus; Amg, amygdala. Landmarks: Cb,
cerebellum; 3V, third ventricle; opt, optic tract. Scale bar, 500
µm.
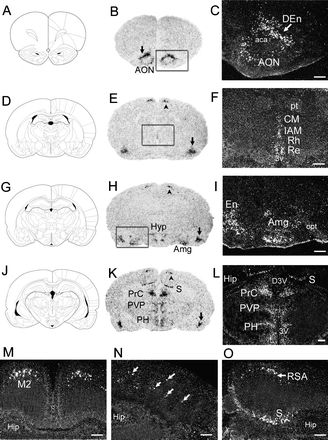
Distribution of NPS Receptor mRNA Expression in Rat Brain.
(A, D, G, and J) Schematic drawings of the sections shown in (B) and
(C) (Bregma, 3.20 mm), (E) and (F) (Bregma -1.80 mm), (H) and (I)
(Bregma -2.80 mm), and (K) and (L) (Bregma -4.52 mm), respectively
(Paxinos and Watson, 1997). (B, E, H, and K) Autoradiograms of NPSR
mRNA expression in coronal rat brain sections. Arrows in (B), (E),
(H), and (K) indicate endopiriform nucleus (En). Arrowheads in (E),
(H), and (K) refer to secondary motor cortex (M2), retrosplenial
agranular cortex (RSA)/M2, and RSA, respectively. (C, F, and I)
Dark-field images of boxed regions in (B), (E), and (H),
respectively. (L) Dark field image of midline thalamic regions of
section (K). (M and N) Dark-field image of cortical regions in
section (E). Arrows in (N) indicate scattered cells expressing NPSR
mRNA in somatosensory cortex. (O) Dark-field image of cortical and
subicular regions in section (K). AON, anterior olfactory nucleus;
DEn, dorsal endopiriform nucleus; CM, central medial thalamic
nucleus; IAM, interanteromedial thalamic nucleus; Rh, rhomboid
thalamic nucleus; Re, reuniens thalamic nucleus; Amg, amygdala; Hyp,
hypothalamus; S, subiculum; Prc, precommissural nucleus; PVP,
paraventricular thalamus nucleus, posterior; PH, posterior
hypothalamus. Landmarks: aca, anterior commissure, anterior part;
pt, paratenial thalamic nuclei; opt, optic tract; D3V, dorsal third
ventricle; 3V, third ventricle; Hip, hippocampus. Scale bar, 500
µm.
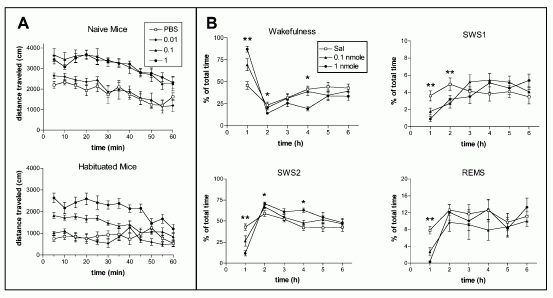
Central Administration of NPS Produces Behavioral Arousal and
Wakefulness. (A) Hyperlocomotion effects of NPS in naive and
habituated mice. Naive mice were new to the test chamber, while
habituated animals were acclimatized for 1 hr prior to the
injection. In naive mice, 0.1 and 1 nmole NPS induce significant
hyperlocomotion (F3,324 = 92.83, p < 0.0001, two-way ANOVA for
repeated measures). The same doses of NPS also produced significant
effects in habituated animals (F3,336 = 135.59, p < 0.0001). (B)
Arousal promoting effects of NPS in rats. NPS increases the amount
of wakefulness and decreases SWS1, SWS2, and REM sleep in rats (n =
8 for each dose). **p < 0.01, 0.1 nmole and 1.0 nmole compared
with saline; *p < 0.01, 1.0 nmole compared with saline (ANOVA
followed by Scheffe's post hoc test).

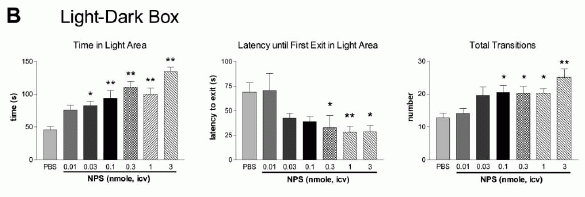
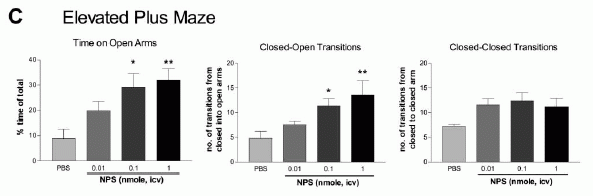
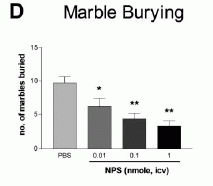
NPS produces dose-dependent anxiolytic-like effects in C57Bl/6 mice exposed to
the open field (A), light-dark box (B), elevated plus maze (C), and
marble burying paradigm (D). Doses and groups: all doses are in
nmole per animal; open field (n = 8 for each dose); light-dark box
(PBS, n = 10; 0.01 nmole, n = 5; 0.03 nmole, n = 5; 0.1 nmole, n =
5; 0.3 nmole, n = 11; 1 nmole, n = 5; 3 nmole, n = 8); elevated plus
maze (n = 5 for all doses); marble burying (PBS and 0.01 nmole, n =
10; 0.1 and 1 nmole, n = 9). **p < 0.01, *p < 0.05 compared to
PBS control, ANOVA followed by Dunnett's test for multiple
comparisons. All data are presented as means ±
SEM.
Tyr10-NPS was labeled with 125I. CHO cells stably
expressing human NPSR were seeded into 24-well plates and cultured
for 48 hr. For saturation binding experiment, [125I]
Tyr10-NPS at concentrations from 4 pM to 1.7 nM were
used. For displacement binding, increasing concentrations of
unlabeled human NPS (1 pM to 3 µM) were used to compete with 0.15 nM
[125I] Tyr10-NPS. Nonspecific binding was determined in
the presence of 1 µM unlabeled human NPS. The binding assay was
carried out as described (Sakurai et al., 1998). In brief, cells
were washed with PBS first and then incubated with radioligand with
or without unlabeled NPS peptide in DMEM medium containing 0.1%
bovine serum albumin at 20°C for 1.5 hr. Cells were washed five
times with cold PBS and lysed with 1 N NaOH. Bound radioactivity was
counted in a MicroBeta liquid scintillation counter (EG&G
Wallac, Gaithersburg, MD) and corrected for counting efficiency.
Data from triplicate incubations were analyzed using
PRISM.
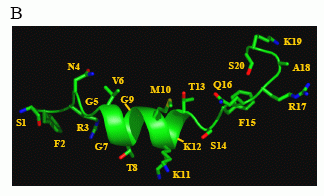
Structural characterization of human NPS by NMR.
B) Putative structural conformation of NPS in the context of
receptor binding, showing an a-helix in the region determined to
contain a nascent helix in the unbound, solubilized peptide.
Bernier
V, et al. J Biol Chem. 2006 Jun 20; [Epub ahead of print]

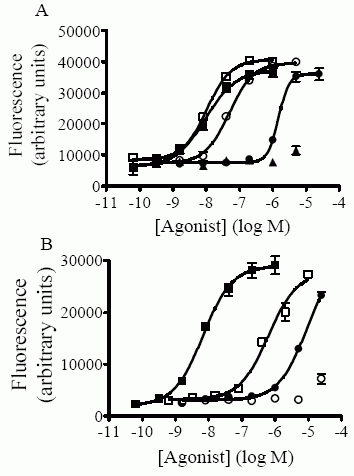
Concentration-response curves in the Ca++-mobilization assay were
generated as described in “Experimental Procedures” and the EC50
values thus obtained are shown in Table II. A) Activation by
C-terminal-truncated NPS peptides. Each point is the mean ± S.D. of
triplicate determinations. Representative experiments of selected
mutant peptides are shown. WT (filled squares);1-13 (empty squares);
1-7 (fille d circles); 1-6 (empty circles); 1-5 (filled triangles).
B) Activation by N-terminal-truncated NPS peptides. Each point is
the mean ± S.D. of triplicate determinations. Representative
experiments are shown. WT (filled squares); 2-20 (empty squares);
3-20 (filled circles); 4-20 (empty circle s). C) Summary of EC50
values obtained for truncated NPS mutant peptides. Each value is the
mean ± S.E.M. of at least three separate determinations for each
peptide and corresponds to values shown in Table II. Peptides for
which EC50 values could not be obtained due to low activity and
absence of a maximal plateau (1-5, 3-20 and 4-20) are shown as
having an EC50 of 2000 nM. Values above bars represent % of maximal
activity (based on WT peptide) obtained for these mutant peptides at
a concentration of 2000 nM.
Bernier V, et al. J Biol Chem. 2006 Jun
20; [Epub ahead of print]
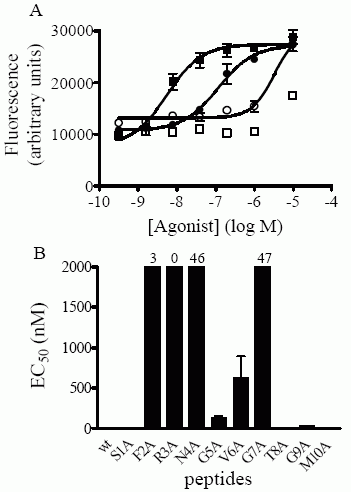
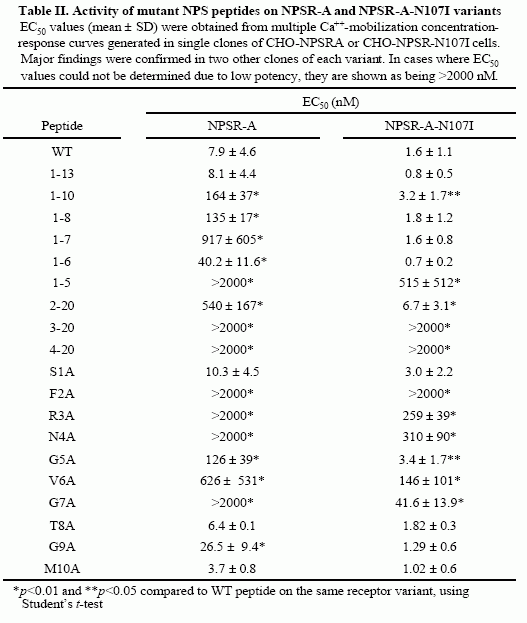
Concentration-response curves
in the Ca++-mobilization
assay were generated as described in “Experimental Procedures” and
the EC50 values thus obtained are shown in Table II. A)
Representative curves of selected mutant peptide. Each point is the
mean ± S.D. of triplicate determinations. WT (filled squares); N4A
(empty squares); V6A (filled circles); G7A (empty circles). C)
Summary of EC50 values obtained for alanine point mutant peptides.
Each value is the mean ± S.E.M. of at least three separate
determinations for each peptide and corresponds to values shown in
Table II. Peptides for which EC50 values could not be obtained due
to low activity and absence of a maximal plateau (F2A, R3A, N4A and
G7A) are shown as having an EC50 of 2000 nM. Values above bars
represent % of maximal activity (based on WT peptide) obtained for
these mutant peptides at a concentration of 2000 nM.
Bernier V, et
al. J Biol Chem. 2006 Jun 20; [Epub ahead of print]
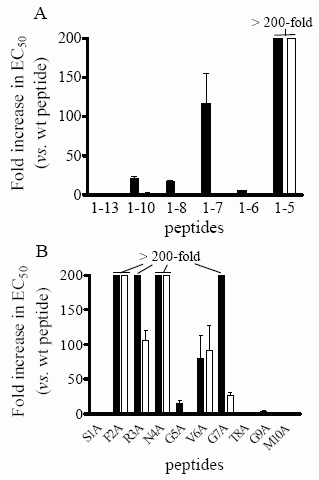
Concentration-response
curves in the Ca++-mobilization assay were generated as described
in “Experimental Procedures” and the EC50 values thus obtained
are shown in Table II. EC50 values of the various mutants were
normalized to those obtained for WT peptide on each variant and are
expressed as fold-increase over WT peptide. Each value is the mean ±
S.E.M. of at least three separate determinations for each peptide.
Peptides for which the fold-increase in EC50 is higher than 200 are
shown as having a 200-fold increase. NPSR-A (filled squares);
N107I-NPSR-A (empty squares). A) Summary of foldincreases in EC50
values for C-terminal-truncated peptides. B) Summary of
fold-increases in EC50 values for alanine point mutant peptides.
Bernier V, et al. J Biol Chem. 2006 Jun 20; [Epub ahead of print]
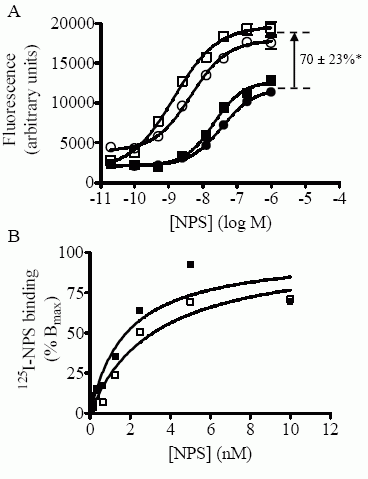
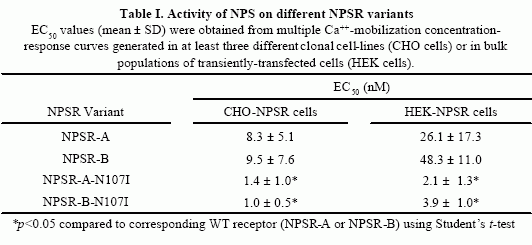
HEK 293 T
cells. A) Concentration-response curves in the Ca++-mobilization
assay were
generated as described in “Experimental Procedures”
and the EC50 values thus obtained are shown in Table I. Each point
is the mean ± S.D. of quadruplicate determinations. A representative
experiment is shown. FLAG-NPSR-A (filled squares); FLAG-NPSR-A-N107I
(empty squares); FLAG-NPSR-B (filled circles); FLAG-NPSR-B-N107I
(empty circles). *p<0.01 using Stduent’s t test. B)
Concentrationdependent binding of 125I-NPS to whole cells expressing
NPSR-A or NPSR-A-N107I variants. Specific binding corresponds to the
difference in binding in the absence and presence of excess
unlabeled NPS (See “Experimental Procedures”). Data is expressed as
percent of calculated Bmax. A representative experiment is shown.
FLAG-NPSR-A (filled squares); FLAG-NPSR-A-N107I (empty squares).
Bernier V, et al. J Biol Chem. 2006 Jun 20; [Epub ahead of print]
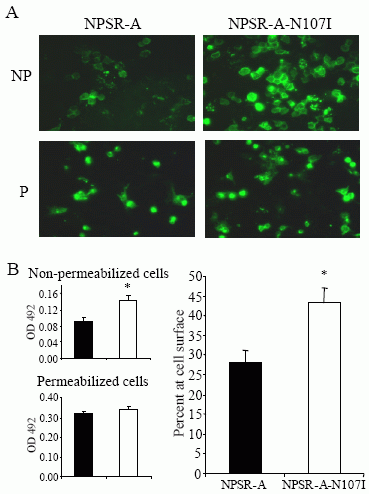
EK 293 T cells. A)
Immunofluorescence microscopy of permeabilized and non-permeabilized
cells
transiently expressing FLAG-NPSR-A and FLAG-NPSR-A-N107I
variants was carried out as described
in “Experimental
Procedures”. “NP” = non permeabilized cells; “P” = permeabilized
cells B) ELISA
determinations on non-permeabilized (upper left
panel) and permeabilized (lower left panel) cells
transiently
expressing FLAG-NPSR-A (filled bars) and FLAG-NPSR-A-N107I (empty
bars) variants
were carried out as described in “Experimental
Procedures”. Right panel: normalized cell surface
expression of
NPSR-A variants, expressed as percent of total receptor expression
(100 x OD492 for nonpermeabilized cells / OD492 for permeabilized
cells). *p<0.01 compared to NPSR-A using Student’s t-test.
Bernier V, et al. J Biol Chem. 2006 Jun 20; [Epub ahead of print]
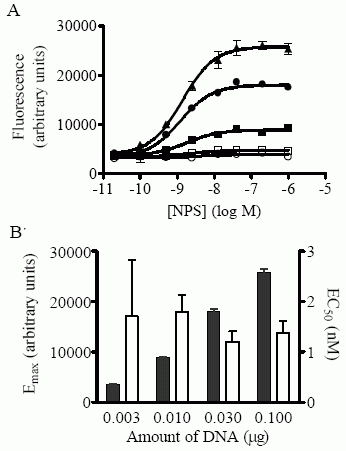
(Ca++-mobilization
assay) are shown in cells transiently transfected with 0 (empty
circles), 0.003 (empty squares), 0.01 (filled squares), 0.03 (filled
circles) and 0.1 (filled triangles) mg of pcDEF3(FLAG-NPSRA- N107I)
vector DNA, supplemented with pcDEF3 vector to maintain a constant
total DNA concentration, as described in “Results”. Each point is
the mean ± S.D. of quadruplicate determinations. A representative
experiment is shown. B) Emax (filled bars) and EC50 (empty bars)
values (± S.E.M.) obtained in transfections using various amounts of
pcDEF3(FLAG-NPSR-A-N107I) DNA, corresponding to the representative
experiment shown in A).
Bernier V, et al. J Biol Chem. 2006 Jun 20;
[Epub ahead of print]
Arousal and anxiety are behavioral responses that involve
complex neurocircuitries and multiple neurochemical components.
Here, we report that a neuropeptide, neuropeptide S (NPS), potently
modulates wakefulness and could also regulate anxiety. NPS acts by
activating its cognate receptor (NPSR) and inducing mobilization of
intracellular Ca2+. The NPSR mRNA is widely distributed in the
brain, including the amygdala and the midline thalamic nuclei.
Central administration of NPS increases locomotor activity in mice
and decreases paradoxical (REM) sleep and slow wave sleep in rats.
NPS was further shown to produce anxiolytic-like effects in mice
exposed to four different stressful paradigms. Interestingly, NPS is
expressed in a previously undefined cluster of cells located between
the locus coeruleus (LC) and Barrington's nucleus. These results
indicate that NPS could be a new modulator of arousal and anxiety.
They also show that the LC region encompasses distinct nuclei
expressing different arousal-promoting
neurotransmitters.
Xu Y.L., et al. Neuron, Vol 43, 487-497, 19 August 2004 (All peptides used in this publication are manufactured by Phoenix Pharmaceuticals)
Many different neuropharmacological agents modulate arousal
and anxiety, yet to date, few endogenous substances have produced
arousal with an anxiolytic effect. In this issue of Neuron, Xu et
al. describe the localization and characterization of a novel
neuropeptide, neuropeptide S (and its cognate receptor), that is
unique in its arousing and anxiolytic-like
properties.
George F. Koob, and Thomas N. Greenwell. Neuron, Vol 43, 487-497, 19 August 2004
Neuropeptide S (NPS) and its receptor (NPSR), are
thought to have a role in asthma pathogenesis; a number of single
nucleotide polymorphisms (SNPs) within NPSR have been shown to be
associated with an increased prevalance of asthma. One such SNP
leads to the missense mutation N107I, which results in an increase
in the potency of NPS for NPSR. In order to gain insight into
structure-function relationships within NPS and NPSR, we first
carried out a limited structural characterization of NPS and
subjected the peptide to extensive mutagenesis studies. Our results
show that the N-terminal third of NPS, in particular residues Phe 2,
Arg 3, Asn 4 and Val 6, are necessary and sufficient for activation
of NPSR. Furthermore, part of a nascent helix within the peptide,
spanning residues 5 through 13, acts as a regulatory region that
inhibits receptor activation. Notably, this inhibition is absent in
the asthma-linked N107I variant of NPSR, suggesting that residue 107
interacts with the aforementioned regulatory region of NPS. While
this interaction may be at the root of the increase in potency
associated with the N107I variant, we show here that the mutation
also causes an increase in cell-surface expression of the mutant
receptor, leading to a concomitant increase in the maximal efficacy
(Emax) of NPS. Our results identify the key residues of NPS involved
in NPSR activation and suggest a molecular basis for the functional
effects of the N107I mutation and for its putative
pathophysiological link with asthma.
Bernier V, et al. J Biol Chem. 2006 Jun 20; [Epub ahead of print]
| |
Binding of NPS Receptor |
| Number of
Samples |
FLIPR (EC50,
nM) |
(IC50,
nM) |
kd
(nM) |
Bmax
(fmol/150000cells) |
| Human NPS |
9.4 +- 3.2 |
0.42 +- 0.12 |
|
|
| Rat NPS |
3.2 +- 1.1 |
|
|
|
| Mouse NPS |
3.0 +- 1.3 |
|
|
|
| Human NPS, 125I-Tyr10 |
6.7 +- 2.4 |
|
0.33+-0.12 |
3.2+-0.4 |

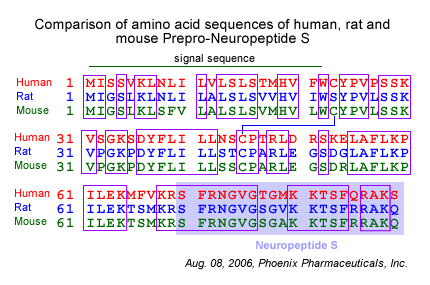
Primary Structures of Neuropeptide S from Human, Chimpanzee,
Rat, Mouse, Dog, and Chicken. Amino acids divergent from the human
sequence are shown in bold type. Sequences were deduced from GenBank
entries BD168686 (human), BD168712 (rat), BD168690 (mouse), BU293859
(chicken), and genome sequencing traces 231487919 (chimpanzee) and
250468833 (dog).
Xu Y.L., et al. Neuron, Vol 43, 487-497, 19 August 2004
|
|
|
%005-71%;%005-72%;%005-77%;%005-78%;%005-79%;%005-80%;%005-81%;%005-83%;%005-84%;%005-85%;%005-86%;%005-87%;%005-88%;%005-89%;%005-90%;%005-91%;%005-92%;%005-93%;%005-94%;%005-95%;%005-96%;%005-97%;%005-98%;%005-99%
|
|
|


